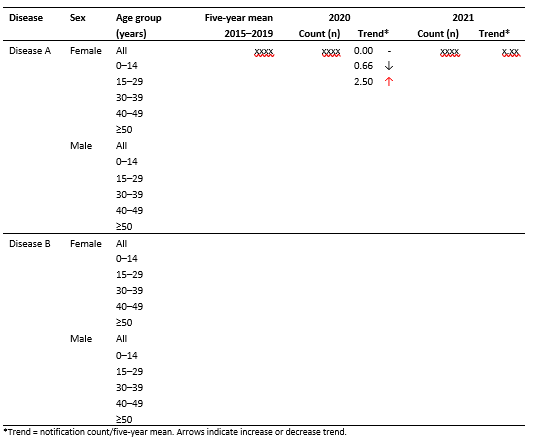
Hello @mdal,
What you are looking to do is definitely feasible within R. Below is an example of how I would tackle the problem. However, there are two options to generalize: you could write a function that takes the years you want to include in the 5-year average calculation and then another to take the years you want to include as counts. Alternatively, you could try using the slider package to calculate 5-year rolling averages for each year.
# loading packages
library(tidyverse)
library(gt)
# creating linelist data
linelist_data <- tibble(
year = sample(
x = 2015:2021,
size = 500,
replace = TRUE
),
disease = sample(
x = c("Disease A", "Disease B"),
size = 500,
replace = TRUE
),
sex = sample(
x = c("Female", "Male"),
size = 500,
replace = TRUE
),
age_group = sample(
x = c("0-14", "15-29", "30-39", "40-49", ">=50"),
size = 500,
replace = TRUE
),
n = rpois(n = 500, lambda = 65)
) |>
mutate(
disease = factor(x = disease, levels = c("Disease A", "Disease B")),
sex = factor(x = sex, levels = c("Female", "Male")),
age_group = factor(age_group, levels = c("0-14", "15-29", "30-39", "40-49", ">=50"))
)
# aggregating linelist data
aggregated_data <- linelist_data |>
count(disease,
year,
sex,
age_group) |>
complete(disease,
year = 2015:2021,
sex,
age_group,
fill = list(n = 0))
# creating average data for 2015 to 2019
averages <- aggregated_data |>
filter(between(year, 2015, 2019)) |>
group_by(disease,
sex,
age_group) |>
summarise(average = mean(n),
.groups = "drop")
# creating counts data for 2020 and 2021 in a wide format
counts <- aggregated_data |>
filter(between(year, 2020, 2021)) |>
pivot_wider(
id_cols = c(disease, sex, age_group),
names_prefix = "count_",
names_from = year,
values_from = n
)
# joining the counts and average data, you can adjust how trend is defined
joined_data <- counts |>
left_join(averages,
by = c("disease", "sex", "age_group")) |>
mutate(
trend_2020 = sign(count_2020 - average),
trend_2021 = sign(count_2021 - average)
) |>
relocate(disease,
sex,
age_group,
average,
count_2020,
trend_2020,
count_2021,
trend_2021)
# using gt to create a nice table - you could look into creating group rows
joined_data |>
gt() |>
cols_label(
disease = "Disease",
sex = "Sex",
age_group = "Age Group",
average = "5 year mean",
count_2020 = "Count",
trend_2020 = "Trend",
count_2021 = "Count",
trend_2021 = "Trend"
) |>
tab_spanner(label = "2020",
columns = c("count_2020", "trend_2020")) |>
tab_spanner(label = "2021",
columns = c("count_2021", "trend_2021"))
Created on 2022-05-19 by the reprex package (v2.0.1)
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.1.3 (2022-03-10)
#> os macOS Big Sur/Monterey 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_CA.UTF-8
#> ctype en_CA.UTF-8
#> tz America/Toronto
#> date 2022-05-19
#> pandoc 2.17.1.1 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.0)
#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.1.0)
#> broom 0.8.0 2022-04-13 [1] CRAN (R 4.1.3)
#> cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.1.0)
#> checkmate 2.1.0 2022-04-21 [1] CRAN (R 4.1.3)
#> cli 3.3.0 2022-04-25 [1] CRAN (R 4.1.3)
#> colorspace 2.0-3 2022-02-21 [1] RSPM (R 4.1.2)
#> crayon 1.5.1 2022-03-26 [1] CRAN (R 4.1.3)
#> DBI 1.1.2 2021-12-20 [1] CRAN (R 4.1.1)
#> dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.1.0)
#> digest 0.6.29 2021-12-01 [1] CRAN (R 4.1.1)
#> dplyr * 1.0.9 2022-04-28 [1] CRAN (R 4.1.3)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
#> evaluate 0.15 2022-02-18 [1] RSPM (R 4.1.2)
#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.1.3)
#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
#> forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.1.0)
#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.1.1)
#> generics 0.1.2 2022-01-31 [1] RSPM (R 4.1.2)
#> ggplot2 * 3.3.6 2022-05-03 [1] CRAN (R 4.1.3)
#> glue 1.6.2 2022-02-24 [1] RSPM (R 4.1.2)
#> gt * 0.5.0 2022-04-21 [1] CRAN (R 4.1.3)
#> gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
#> haven 2.5.0 2022-04-15 [1] CRAN (R 4.1.3)
#> highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
#> hms 1.1.1 2021-09-26 [1] CRAN (R 4.1.1)
#> htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
#> httr 1.4.3 2022-05-04 [1] CRAN (R 4.1.2)
#> jsonlite 1.8.0 2022-02-22 [1] RSPM (R 4.1.2)
#> knitr 1.39 2022-04-26 [1] CRAN (R 4.1.3)
#> lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.1)
#> lubridate 1.8.0 2021-10-07 [1] CRAN (R 4.1.1)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.1.3)
#> modelr 0.1.8 2020-05-19 [1] CRAN (R 4.1.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
#> pillar 1.7.0 2022-02-01 [1] RSPM (R 4.1.2)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
#> purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
#> R.cache 0.15.0 2021-04-30 [1] CRAN (R 4.1.0)
#> R.methodsS3 1.8.1 2020-08-26 [1] CRAN (R 4.1.0)
#> R.oo 1.24.0 2020-08-26 [1] CRAN (R 4.1.0)
#> R.utils 2.11.0 2021-09-26 [1] CRAN (R 4.1.1)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
#> readr * 2.1.2 2022-01-30 [1] RSPM (R 4.1.2)
#> readxl 1.4.0 2022-03-28 [1] CRAN (R 4.1.3)
#> reprex 2.0.1 2021-08-05 [1] CRAN (R 4.1.0)
#> rlang 1.0.2 2022-03-04 [1] CRAN (R 4.1.2)
#> rmarkdown 2.14 2022-04-25 [1] CRAN (R 4.1.3)
#> rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
#> rvest 1.0.2 2021-10-16 [1] CRAN (R 4.1.1)
#> sass 0.4.1 2022-03-23 [1] CRAN (R 4.1.2)
#> scales 1.2.0 2022-04-13 [1] CRAN (R 4.1.3)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.1.1)
#> stringi 1.7.6 2021-11-29 [1] CRAN (R 4.1.1)
#> stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
#> styler 1.7.0 2022-03-13 [1] CRAN (R 4.1.2)
#> tibble * 3.1.7 2022-05-03 [1] CRAN (R 4.1.3)
#> tidyr * 1.2.0 2022-02-01 [1] RSPM (R 4.1.2)
#> tidyselect 1.1.2 2022-02-21 [1] RSPM (R 4.1.2)
#> tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.1.0)
#> tzdb 0.3.0 2022-03-28 [1] CRAN (R 4.1.3)
#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
#> vctrs 0.4.1 2022-04-13 [1] CRAN (R 4.1.3)
#> withr 2.5.0 2022-03-03 [1] RSPM (R 4.1.2)
#> xfun 0.31 2022-05-10 [1] CRAN (R 4.1.3)
#> xml2 1.3.3 2021-11-30 [1] CRAN (R 4.1.1)
#> yaml 2.3.5 2022-02-21 [1] RSPM (R 4.1.2)
#>
#> [1] /Users/timothychisamore/Library/R/x86_64/4.1/library
#> [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────
All the best,
Tim