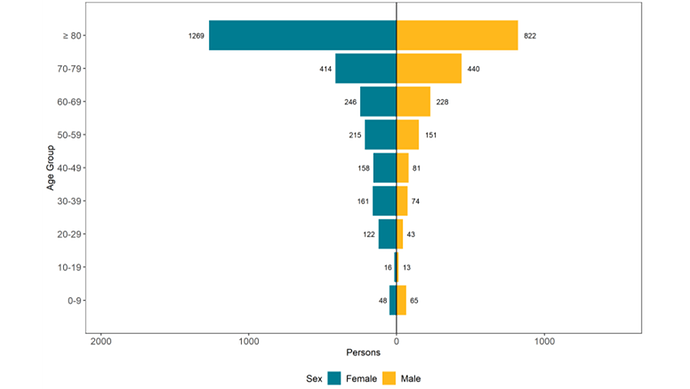Hello,
For my age-sex pyramid I want to display the counts next to the bars they correspond to but I’m not having much luck doing so.
My reprex is below, TIA!
pacman::p_load(epikit, apyramid, tidyverse)
pyramid_data <- data.frame(
stringsAsFactors = FALSE,
AGE_IN_YEARS = c(87L,75L,66L,1L,60L,65L,0L,
82L,37L,55L,83L,92L,95L,90L,86L,94L,80L,82L,
82L,64L,41L,76L,71L,61L,63L,0L,85L,70L,58L,87L,
65L,37L,66L,55L,50L,75L,15L,70L,95L,64L,87L,
92L,79L,81L,84L,92L,85L,90L,82L,66L,71L,1L,71L,
79L,79L,78L,0L,2L,85L,68L,84L,85L,79L,93L,
79L,78L,76L,70L,89L,61L,84L,89L,0L,80L,95L,71L,
70L,84L,61L,89L,59L,21L,35L,68L,0L,83L,95L,85L,
92L,76L,78L,58L,76L,87L,79L,80L,7L,32L,77L,
77L),
PATIENT_SEX = c("Male","Female","Female",
"Female","Female","Male","Female","Male","Female",
"Female","Female","Male","Female","Male","Female",
"Male","Male","Female","Male","Female","Female","Male",
"Female","Female","Female","Male","Male","Male",
"Male","Male","Female","Female","Female","Male",
"Female","Male","Male","Male","Male","Female","Male",
"Male","Male","Male","Female","Female","Female",
"Female","Female","Male","Female","Female","Male","Male",
"Male","Female","Male","Female","Female","Female",
"Male","Male","Female","Female","Male","Male",
"Female","Female","Female","Male","Female","Female",
"Female","Female","Female","Male","Female","Male",
"Male","Female","Female","Female","Female","Female",
"Male","Male","Female","Male","Female","Male","Female",
"Female","Female","Male","Female","Male","Female",
"Male","Female","Female"),
age_cat = as.factor(c("80+",
"70-79","60-69","0-9","60-69","60-69","0-9",
"80+","30-39","50-59","80+","80+","80+",
"80+","80+","80+","80+","80+","80+","60-69",
"40-49","70-79","70-79","60-69","60-69",
"0-9","80+","70-79","50-59","80+","60-69",
"30-39","60-69","50-59","50-59","70-79",
"10-19","70-79","80+","60-69","80+","80+",
"70-79","80+","80+","80+","80+","80+",
"80+","60-69","70-79","0-9","70-79","70-79",
"70-79","70-79","0-9","0-9","80+","60-69",
"80+","80+","70-79","80+","70-79","70-79",
"70-79","70-79","80+","60-69","80+",
"80+","0-9","80+","80+","70-79","70-79","80+",
"60-69","80+","50-59","20-29","30-39",
"60-69","0-9","80+","80+","80+","80+",
"70-79","70-79","50-59","70-79","80+","70-79",
"80+","0-9","30-39","70-79","70-79"))
)
palette <- c("#007C91", "#FFB81C")
age_sex_pyramid <- age_pyramid(
data = pyramid_data,
age_group = age_cat,
split_by = PATIENT_SEX,
proportional = TRUE,
show_midpoint = FALSE
) +
geom_col(aes(fill = PATIENT_SEX), color = "white", position = "identity") +
theme_classic() +
scale_fill_manual(values = palette) +
labs(
title = "Age and sex of cases, for the past 4 weeks",
x = "Age Group",
y = "Proportion of all cases",
fill = ""
) +
theme(
plot.title = element_text(color = "#007C91", size = 16, face = "bold", hjust = 0.5)
)
Created on 2024-03-13 with reprex v2.1.0
Session info
sessionInfo()
#> R version 4.3.2 (2023-10-31 ucrt)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 19045)
#>
#> Matrix products: default
#>
#>
#> locale:
#> [1] LC_COLLATE=English_United Kingdom.utf8
#> [2] LC_CTYPE=English_United Kingdom.utf8
#> [3] LC_MONETARY=English_United Kingdom.utf8
#> [4] LC_NUMERIC=C
#> [5] LC_TIME=English_United Kingdom.utf8
#>
#> time zone: Europe/London
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] digest_0.6.34 fastmap_1.1.1 xfun_0.42 glue_1.7.0
#> [5] knitr_1.45 htmltools_0.5.7 rmarkdown_2.25 lifecycle_1.0.4
#> [9] cli_3.6.2 reprex_2.1.0 withr_3.0.0 compiler_4.3.2
#> [13] rstudioapi_0.15.0 tools_4.3.2 evaluate_0.23 yaml_2.3.8
#> [17] rlang_1.1.3 fs_1.6.3