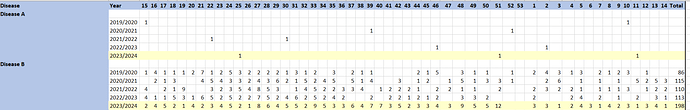
Hello Donna,
Here is how I would approach the problem, note that it is pretty in depth as I mentioned since you need to check whether you are currently in the first epiweek when creating the table:
# loading packages
library(tidyverse)
# creating fake data
fake_data <-
tibble(
date = sample(
x = seq.Date(
from = as.Date("2019-01-01"),
to = today(),
by = "day"
),
size = 10000,
replace = TRUE
),
disease = sample(
x = c("Disease A", "Disease B"),
size = 10000,
replace = TRUE
)
)
# shaping data
fake_data_shaped <- fake_data |>
mutate(
date = ymd(date),
epiyear = epiyear(date),
epiweek = epiweek(date)
) |>
count(disease,
epiyear,
epiweek) |>
# filling in missing combinations of disease, epiyear, and epiweek
complete(
disease = unique(disease),
epiyear = seq(from = min(epiyear), to = max(epiyear)),
epiweek = seq(from = 1, to = 53),
fill = list(n = 0)
) |>
# creating a label for combined years and a variable to sort the data
mutate(year_label = case_when(
epiweek(today()) != 1 ~ paste(epiyear, epiyear + 1, sep = "/"),
.default = as.character(epiyear)
),
sort = (((
epiweek - epiweek(today()) - 1
) %% 53) + 1)) |>
# sorting the data
arrange(disease, year_label, sort) |>
select(-c(sort, epiyear)) |>
# transposing the data into a wide format
pivot_wider(names_from = epiweek,
names_sort = FALSE,
values_from = n,
values_fill = 0)
# displaying the data
fake_data_shaped |>
slice_head(n = 5)
#> # A tibble: 5 × 55
#> disease year_label `18` `19` `20` `21` `22` `23` `24` `25` `26` `27`
#> <chr> <chr> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 Diseas… 2019/2020 13 14 13 16 16 12 15 13 12 23
#> 2 Diseas… 2020/2021 13 15 13 17 15 22 15 18 21 21
#> 3 Diseas… 2021/2022 17 20 13 20 20 23 17 19 16 24
#> 4 Diseas… 2022/2023 19 13 17 12 18 18 20 20 17 14
#> 5 Diseas… 2023/2024 17 21 14 12 16 14 13 15 23 14
#> # ℹ 43 more variables: `28` <int>, `29` <int>, `30` <int>, `31` <int>,
#> # `32` <int>, `33` <int>, `34` <int>, `35` <int>, `36` <int>, `37` <int>,
#> # `38` <int>, `39` <int>, `40` <int>, `41` <int>, `42` <int>, `43` <int>,
#> # `44` <int>, `45` <int>, `46` <int>, `47` <int>, `48` <int>, `49` <int>,
#> # `50` <int>, `51` <int>, `52` <int>, `53` <int>, `1` <int>, `2` <int>,
#> # `3` <int>, `4` <int>, `5` <int>, `6` <int>, `7` <int>, `8` <int>,
#> # `9` <int>, `10` <int>, `11` <int>, `12` <int>, `13` <int>, `14` <int>, …
Created on 2024-04-26 with reprex v2.1.0
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.3.3 (2024-02-29)
#> os macOS Sonoma 14.4.1
#> system x86_64, darwin20
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz America/Toronto
#> date 2024-04-26
#> pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.0)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
#> digest 0.6.35 2024-03-11 [1] RSPM (R 4.3.0)
#> dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.0)
#> evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.0)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.0)
#> fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
#> fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
#> ggplot2 * 3.5.0 2024-02-23 [1] RSPM (R 4.3.0)
#> glue 1.7.0 2024-01-09 [1] RSPM (R 4.3.0)
#> gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
#> htmltools 0.5.8.1 2024-04-04 [1] RSPM (R 4.3.0)
#> knitr 1.46 2024-04-06 [1] RSPM (R 4.3.0)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.0)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
#> munsell 0.5.1 2024-04-01 [1] RSPM (R 4.3.0)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
#> R.cache 0.16.0 2022-07-21 [1] CRAN (R 4.3.0)
#> R.methodsS3 1.8.2 2022-06-13 [1] CRAN (R 4.3.0)
#> R.oo 1.26.0 2024-01-24 [1] RSPM (R 4.3.0)
#> R.utils 2.12.3 2023-11-18 [1] CRAN (R 4.3.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
#> readr * 2.1.5 2024-01-10 [1] RSPM (R 4.3.0)
#> reprex 2.1.0 2024-01-11 [1] RSPM (R 4.3.0)
#> rlang 1.1.3 2024-01-10 [1] RSPM (R 4.3.0)
#> rmarkdown 2.26 2024-03-05 [1] RSPM (R 4.3.0)
#> rstudioapi 0.16.0 2024-03-24 [1] RSPM (R 4.3.0)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
#> stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.0)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.0)
#> styler 1.10.3 2024-04-07 [1] RSPM (R 4.3.0)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
#> tidyr * 1.3.1 2024-01-24 [1] RSPM (R 4.3.0)
#> tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.3.0)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
#> timechange 0.3.0 2024-01-18 [1] RSPM (R 4.3.0)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.0)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.0)
#> withr 3.0.0 2024-01-16 [1] RSPM (R 4.3.0)
#> xfun 0.43 2024-03-25 [1] RSPM (R 4.3.0)
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.0)
#>
#> [1] /Users/timothychisamore/Library/R/x86_64/4.3/library
#> [2] /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────
With this, you can then calculate row sums to get your totals and you can rearrange the variables as you see fit.
All the best,
Tim